ALGORITHM
The data needs to be extracted and analysed before it is used for pre processing.We extracted data from various antimicrobial databases such as LAMP,CAMP,APD,PenBase and EROP.We made sure that the sequences selected for positive dataset belonged to family Pisces or Crustasea.The negative dataset was created using mitochondrial peptides obtained from fish.
Before using machine learning algorithm for training and testing, the biological sequences need to be converted to format suitable for input to computer system (arff).For this we calculated total of 35 parameters using BioPerl out of which, class was dependent feature.The other 20 features represented the amino acid composition (AAC) and the other 13 features (viz.Amino acid number,Number of negative amino acids
Number of positive amino acids, Molecular weight, Theoretical pI ,Number of carbon atoms,Number of hydrogen atoms,Number of nitrogen atoms,Number of oxygen atoms, Number of sulphur atoms, Half life, Instability Index, Stability class,stability index,Aliphatic_index, Gravy) were the physic-chemical parameters.
For both Support vector machine (SVM) technique and Artificial Neural Network technique we used WEKA software on command line.
Support Vector Machine technique(SVM) using SMO function
Support vector machines (SVMs) are supervised learning models with associated learning algorithms that analyze data and recognize patterns, used for classification and regression analysis.An SVM model represents examples as points in sapce which are mapped so that seperate categories are divided by a clear gap. New examples are then predicted on the basis on which side of the gap they fall on. SVMs can efficiently perform a non-linear classification using the kernel functions, implicitly mapping their inputs into high-dimensional feature spaces.
SMO(Sequential Minimal Optimization) algorithm was used in the weka software to built a model for future predictions. It solves the SVM QP(Quadratic Programming) problem by involving two Lagrange multipliers. We took a binary classification problem with a dataset (x1, y1), ..., (xn, yn), where xi is an input vector and yi= {0, 1} is a binary label corresponding to it.For prediction of antimicrobial property, the input vector dimension is 35. A soft-margin support vector machine is trained by solving a quadratic programming problem, which is expressed in the dual form as follows:
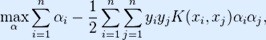
subject to:
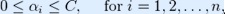
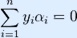
where C is an SVM hyperparameter and K(xi, xj) is the kernel function, both to be supplied by the user; and the variables  are Lagrange multipliers.
are Lagrange multipliers.
SMO breaks this problem into a series of smallest possible sub-problems, which are then solved analytically. Because of the linear equality constraint involving the Lagrange multipliers
 , the smallest possible problem involves two such multipliers. Then, for any two multipliers
, the smallest possible problem involves two such multipliers. Then, for any two multipliers
 and
and  , the constraints are reduced to:
, the constraints are reduced to:
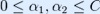
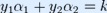
and this reduced problem can be solved analytically: one needs to find a minimum of a one-dimensional quadratic function. k is the negative of the sum over the rest of terms in the equality constraint, which is fixed in each iteration.
The algorithm proceeds as follows:
1. Find a Lagrange multiplier
 that violates the Karush–Kuhn–Tucker (KKT) conditions for the optimization problem.
that violates the Karush–Kuhn–Tucker (KKT) conditions for the optimization problem.
2. Pick a second multiplier  and optimize the pair
and optimize the pair  .
3. Repeat steps 1 and 2 until convergence.
4. When all the Lagrange multipliers satisfy the KKT conditions (within a user-defined tolerance), the problem has been solved. Although this algorithm is guaranteed to converge, heuristics are used to choose the pair of multipliers so as to accelerate the rate of convergence.
.
3. Repeat steps 1 and 2 until convergence.
4. When all the Lagrange multipliers satisfy the KKT conditions (within a user-defined tolerance), the problem has been solved. Although this algorithm is guaranteed to converge, heuristics are used to choose the pair of multipliers so as to accelerate the rate of convergence.
The choice of the proper kernel function is an important issue for SVM training because the power of SVM comes from the kernel representation that allows the nonlinear mapping of input space to a higher dimensional feature space.The kernel functions that we used are:
1. 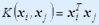 (Linear SVM)
(Linear SVM)
2. 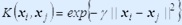 (Radial Basis function Kernel)
(Radial Basis function Kernel)
Support Vector Machine technique(SVM) using Multilayer Perceptron function
A multilayer perceptron (MLP) is a feedforward artificial neural network model;i.e data flows in one direction from input to output layer,with one or more layers between input and output layer. In MLP,each node is a neuron (or processing element) with a nonlinear activation function, except for the input nodes. It uses Backpropagation to train the network.
Activation Function
The activation function of the MLP is a Sigmoid function:

It determines the new level of activation based on the effective input and the current activation. Sigmoid function reduces the computational burden for training.
Backpropagation
The Backpropagation algorithm is used to learn the weights of a multilayer neural network with
a fixed architecture. It performs gradient descent to try to minimize the sum squared error between
the network’s output values and the given target values.
Backpropagation Algorithm can be divided into two phases:
1. Propagation
Steps Involved:
i. Forward Propagation of Trining pattern input is done through the neural network to generate output activations.
ii. Backward Propagation of the output activations is done using the training pattern target in order to generate deltas of all the outputs and hidden nuerons.
2. Weight Update
Steps Involved:
i. Multiplication of output delta and input activation to generate the weight gradient.
ii. Subtraction of a percentage of the gradient from the weight.
This Percentage is called the LEARNING RATE and it influences the speed and quality of learning.
Gradient Descent is used to find the change in weight. Sign of the gradient of weight indicates the increase in error, so wieght update is in the opposite direction.
These steps are repeated until the performance of the network is satisfactory.
Except for when the class is numeric in which case the the output nodes become unthresholded linear units, the nodes in this network are all sigmoid.
Cross Validation
The 5-fold cross validation was used to validate all the models.In 5- fold cross validaton, the original sample is randomly partitioned into 5 equal size Sets(ie. they contained approximately equal number of peptides).A single set is retained as the validation data for testing the model, and the remaining 4 samples are used as training data. The cross-validation process is repeated 5 times ,with each of the 5 sets used as the validation data.The average of results is then taken to make a final estimation.