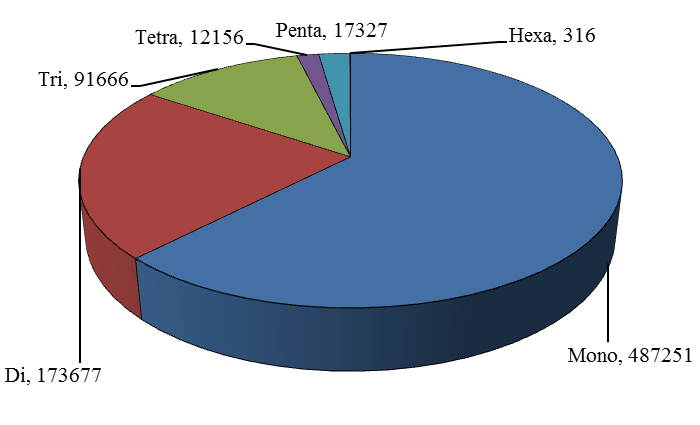
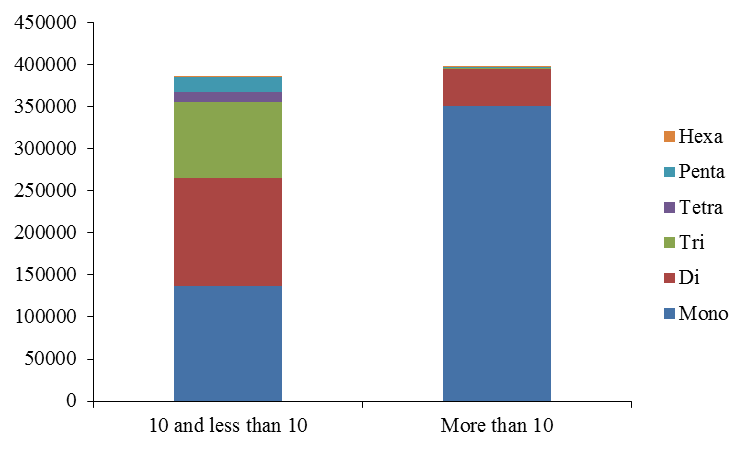The first genome sequence of domestic goat has been completed by integrating next-generation sequencing (NGS) and whole-genome mapping (WGM) technologies. The goat genome is the first reference genome for small ruminant animals and may help to advance the understanding of distinct ruminants' genomic features from non-ruminant species. All the (29 + X) chromosomes were chopped into manageable range of 50000 bp using PERL script. These fragments were given to MIcroSAtellite identification tool (MISA) to identify and find the location of perfect and compound microsatellites for all the chromosomes of goat genome. The SSR numbers, motifs, repeat number, length of the repeat, size of the repeat, repeat type, GC content, start and end position of the repeat and SSR sequence has been compiled. Chromosome wise description of the SSRs have been tabulated as well as represented as graphs. The mono, di, tri, tetra, penta and hexa type of microsatellites have been plotted to show the abundance of the type in chromosomes. In addition, facility to locate the SSRs on chromosomes based on the position number is incorporated which may be very useful to the biotechnologists. The tabulation of simple versus compound shows the proportion of the same. The data can be available on request
Table 1. Description of Goat genome
Table 2. Description of Microsatellite types in Goat genome
Table 3. Motif wise distribution of Microsatellites in Goat genome
27670 9689 5388 711 1150 20 4769 47310 16720 9058 1112 1952 30 7690 37722 13846 7696 1068 1552 22 6384 20487 7276 3924 465 693 9 3415 18880 6687 3596 465 723 8 3049 20432 7155 4087 508 873 17 3544 18531 6435 3491 421 614 12 3007 19080 6849 3929 521 708 14 3290 16245 5866 3114 334 634 9 2590 17070 5773 3293 365 511 7 2681 18295 6369 3285 461 610 7 3112 16771 5593 2787 428 601 9 2920 14501 4914 2480 361 386 9 2288 16620 5997 3149 453 618 17 2938 13358 4793 2699 363 495 6 2357 13412 4869 2509 348 477 9 2304 13288 4755 2355 310 477 9 2350 10635 3958 1821 266 253 4 1832 11352 3900 1802 270 258 9 1891 12510 4650 2424 320 487 9 2273 11497 4252 2082 286 358 9 1981 10473 3442 1734 235 274 7 1710 9111 3056 1594 184 248 8 1600 11321 4024 2033 278 362 12 1885 7265 2607 1203 198 177 3 1246 8770 3098 1623 223 296 7 1569 8613 3056 1442 187 219 4 1413 7164 2664 1479 161 243 5 1260 7841 3103 1501 223 244 3 1475 21027 8281 4088 631 834 22 3994 Table 4. Motif wise distribution of Microsatellites in Goat chromosomes
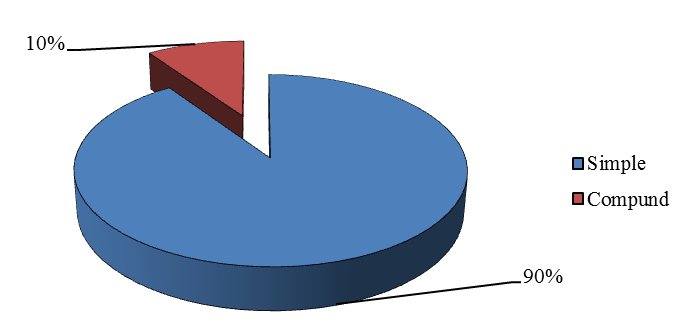
Figure 1. Distribution of SSR marker type
Figure 2. Distribution of repeat types
Figure 3. Distribution of repeat numbers