Input
The input sequences must be supplied in FASTA format. Each sequence should contains only standard amino acid residues in a single letter code format i.e., A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y. An example dataset having 2 protein sequences is given below. If sequences with non-standard residues are supplied, then also prediction will be made only for the sequences having standard residues.
>GAO28253.1 5-formyltetrahydrofolate cyclo-ligase [Geofilum rubicundum JCM 15548]
MRKMEMIQKKKALRKHITYLKSVVPLLQMQEESRHVVAAIEALAVFKQARTVLAYWPMFKEL
DLSSLLNKWQAEKVFLLPVVQGDSLEIRRFQGETSLAPGPSFGIMEPVGPAFNAFGDIDLVL
VPGVAFDEEGRRIGHGKAYYDRLLPQLTKAFKIGVGFSFQLVEDVPSEAHDVRLDLVVVPPM
KPKDIGK
>GAO28252.1 GldB protein [Geofilum rubicundum JCM 15548]
MKLVFVSSIVTLLLMTMACSSGSKAPDVSHVDVDFELIPFYEDLFAIHPDSFAGEAEALKAK
YGQYLEAYSLGVIAAGSTEDEDFVENMQFFLSYEPNQEVLDTCRLVFGDTAPLEEELESAFK
PETPLASLMQMNDGHAFFRSARYQP
Out put
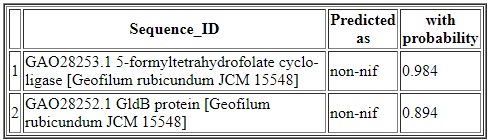
The output (results) are displayed in a tabular format with four columns. The first column represents the serial number, second column represents the sequence identifier, third column represents the types of predicted nif or non-nif and the fourth column represents the probabilities with which they are predicted in the respective classes. For example, the two sequences are predicted as non-nif with probabilities 0.984 and 0.894 respectively. While prediction with proteome-wide data, only the top three predted sequences (with higher probabilities) for each category of nif is displayed. However, the complete result can be downloded from the link download complete result file.