Analysis
The pigeon pea genome was published in Nature Biotechnology in November 2011. The genome was sequenced primarily with Illumina short reads, although assembly was assisted by a number of BAC send sequences produced using traditional Sanger-sequencing long reads. The assembly contains 606 megabases of sequence, a little under three quarters of the estimated total genome size of 833 megabases, and includes an estimated 48,680 genes. While the pidgeon pea genome is made up of 11 chromosomes, the current assembly consists of ~7,000 super scaffolds. Copy of data used in database development and analysis work is available on request or Downloaded from: http://www.icrisat.org/gt-bt/iipg/Genome_Manuscript.html
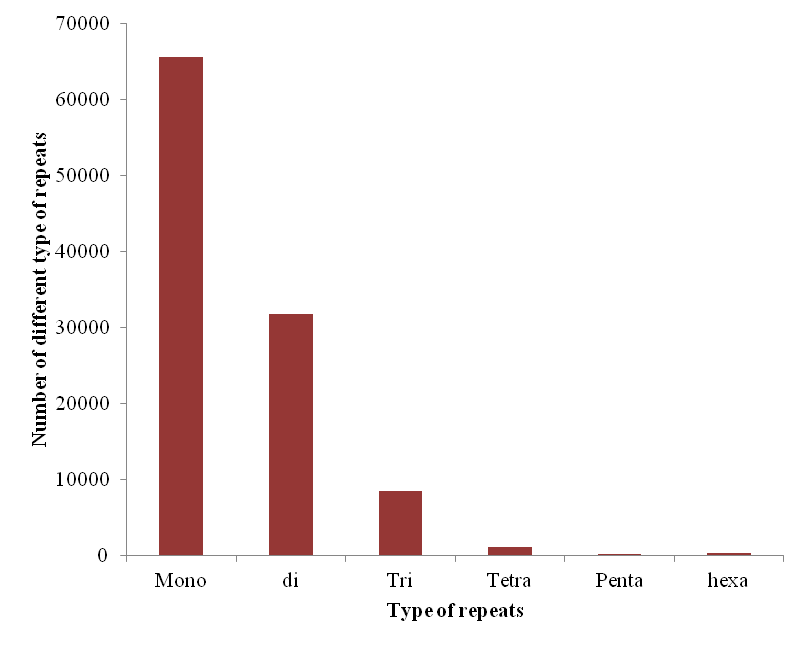
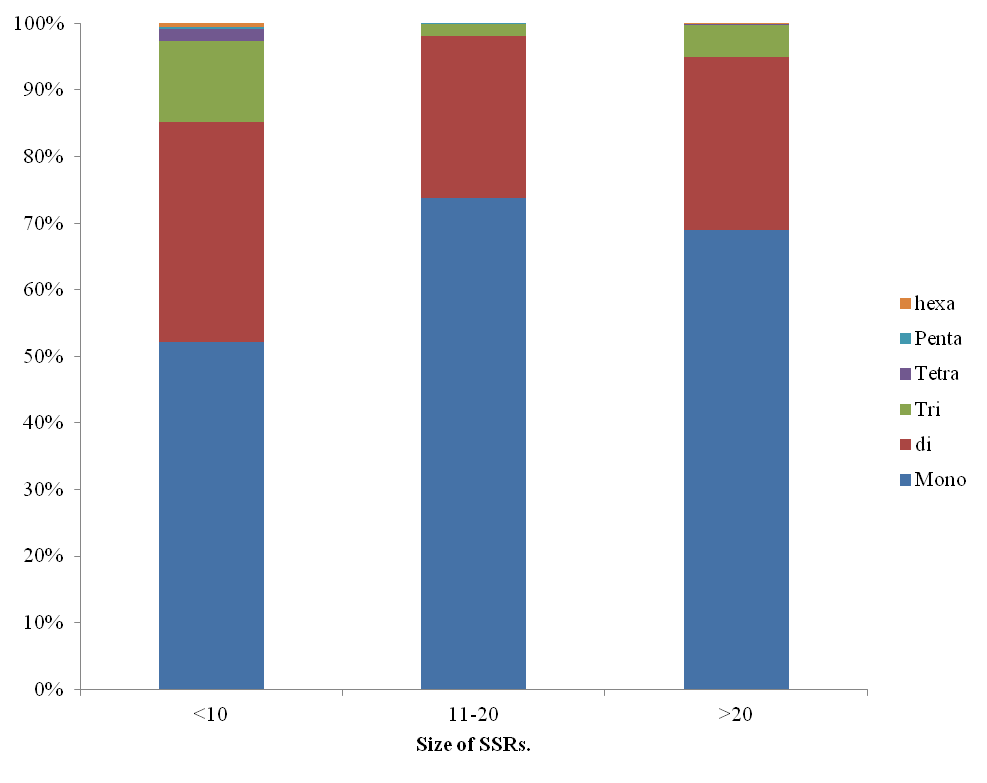
All the 11 chromosomes were chopped into manageable range of 50000 bp using PERL script. These fragments were given to MIcroSAtellite identification tool (MISA) to identify and find the location of perfect and compound microsatellites for all the chromosomes of Pigeonpea genome. The SSR numbers, motifs, repeat number, length of the repeat, size of the repeat, repeat type, GC content, start and end position of the repeat and SSR sequence has been compiled. Chromosome wise description of the SSRs have been tabulated as well as represented as graphs. The mono, di, tri, tetra, penta and hexa type of microsatellites have been plotted to show the abundance of the type in chromosomes.In addition, facility to locate the SSRs on chromosomes based on the position number is incorporated which may be very useful to the biotechnologists. The tabulation of simple versus compound shows the proportion of the same.
Table 1. Description of PigeonPea genome
Table 2. Description of Microsatellite types in PigeonPea genome
Table 3. Motif wise description of Microsatellites in PigeonPea genome
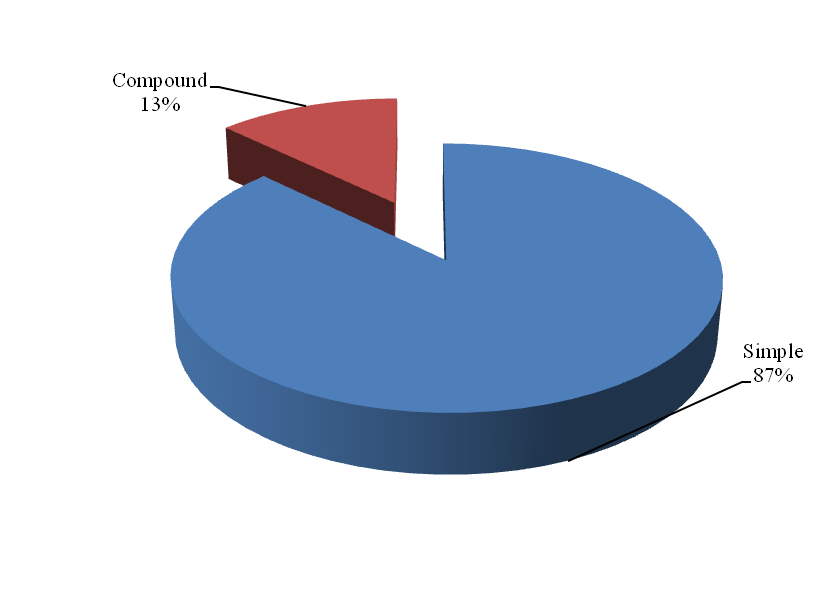 Figure 1. Distribution of SSR marker type
Figure 1. Distribution of SSR marker type
Figure 2. Distribution of repeat types
Figure 3. Distribution of repeat kinds
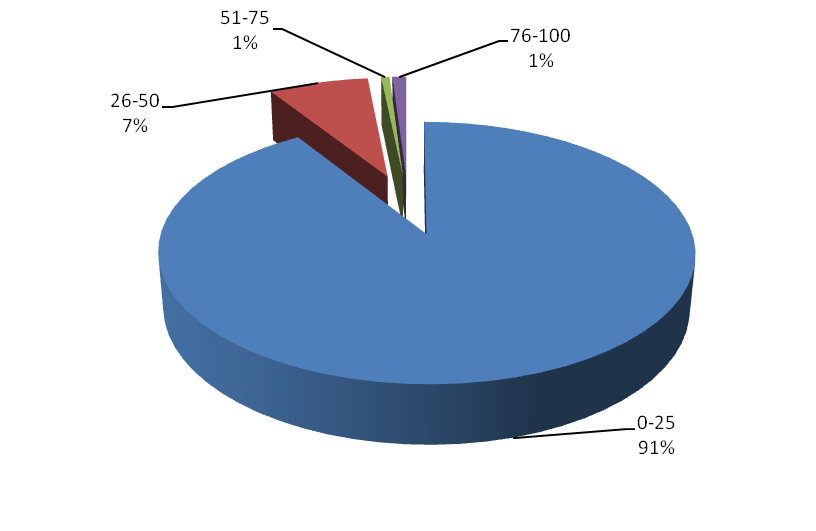
Figure 4. Distribution based on GC Content