Tutorial
PipeMicrodb link: http://cabindb.iasri.res.in/pigeonpea/
Pigeonpea MicroSatellite database (PipeMicrodb) has following tabs:
Home, About, Database, Analysis, Tutorial, Team, Links and Contact.
The “Home” tab describes the database in general, its importance to agricultural biotechnologists and breeders.
The “About” tab gives the description of pigeonpea, its sequencing carried out and the application of microsatellite markers etc.

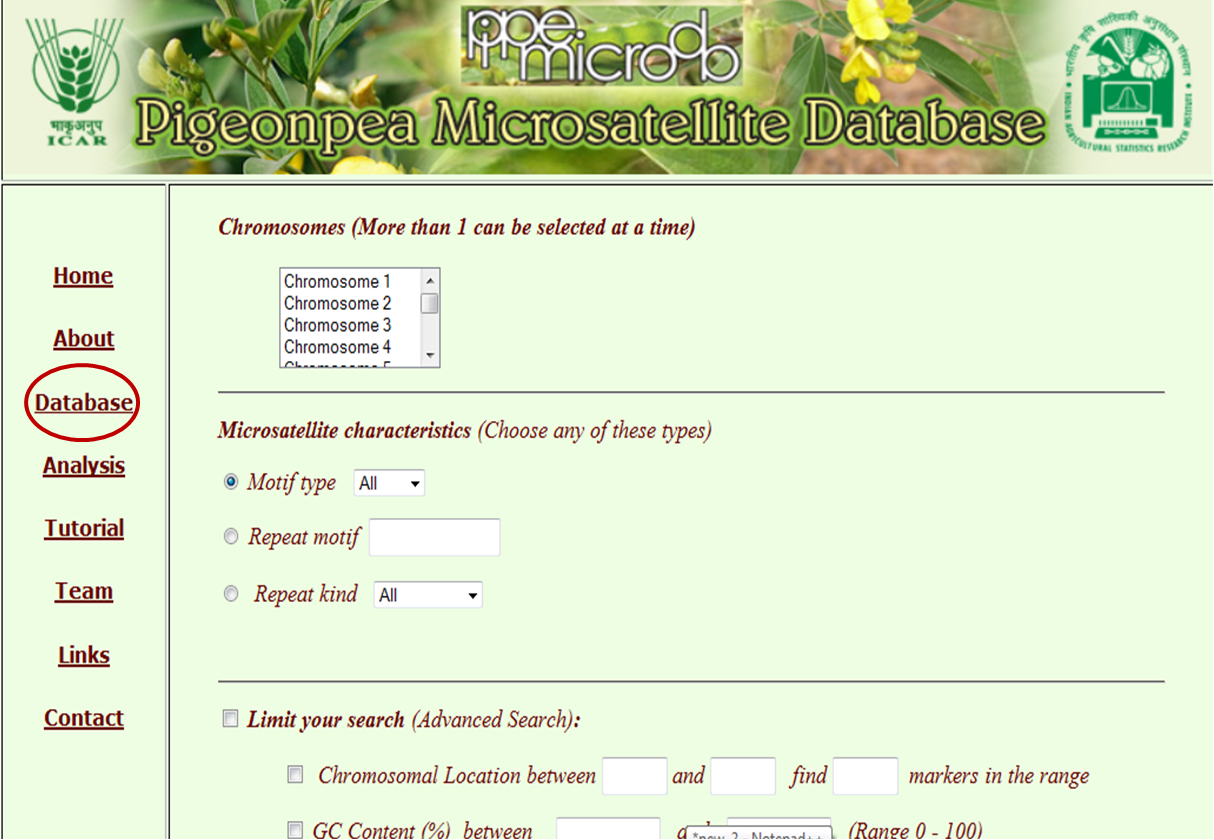
Database query search
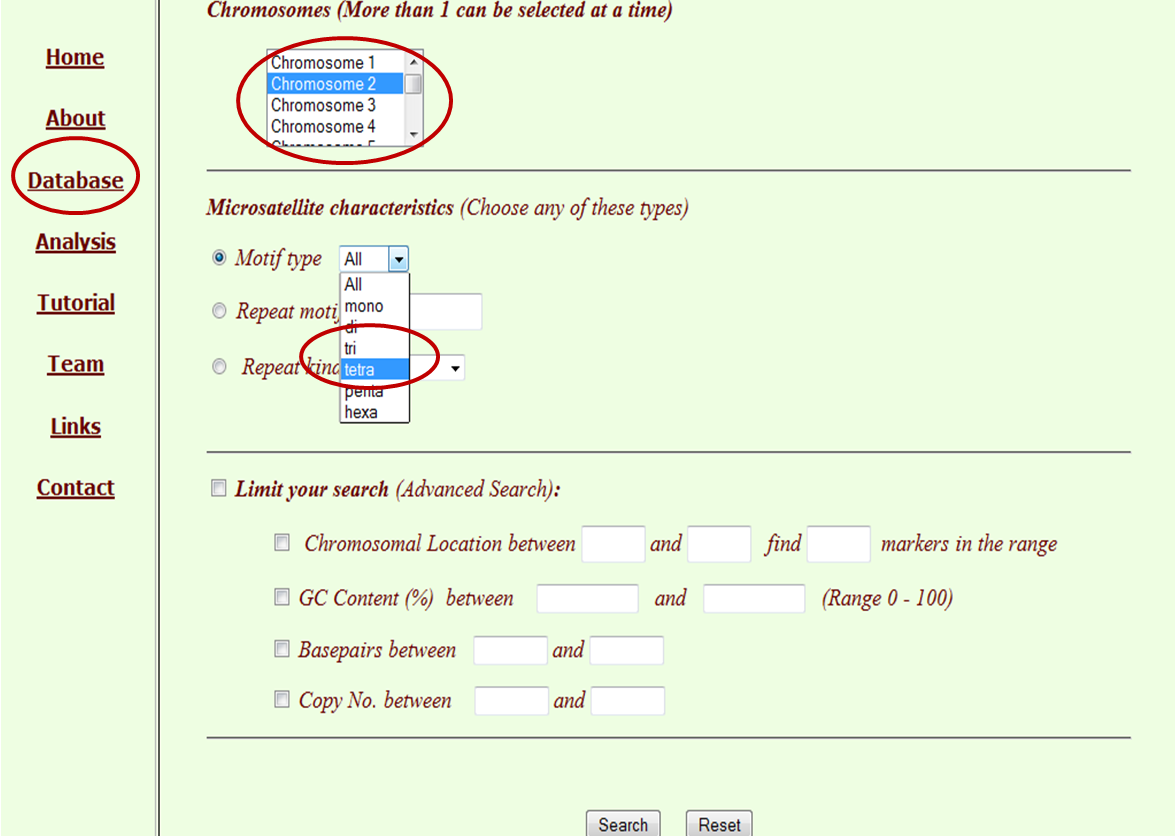
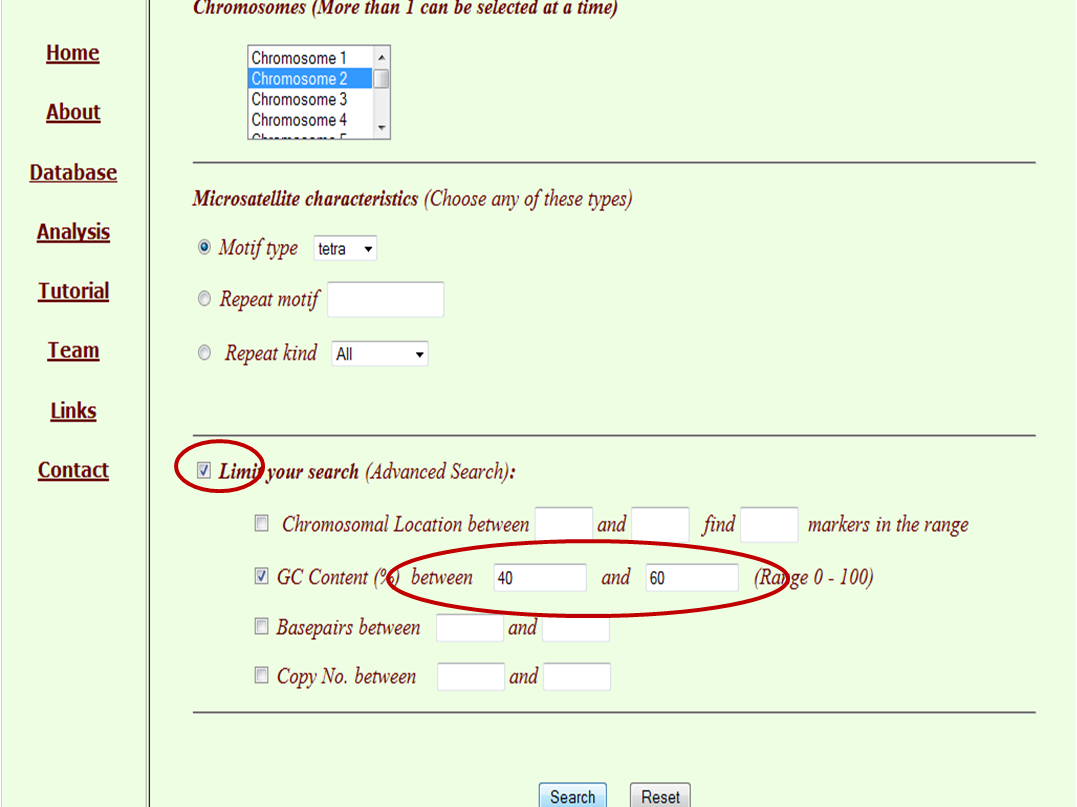
The major search for the database can be made by clicking through “Database”, which is the main tab of this database, where the user has customised options for database search. These searches may be made based on chromosome (multiple selection of chromosome as well as whole genome) and microsatellite characterstics. Microsatellite characterstics include motif type (mono, di, tri, tetra, penta and hexa), repeat type (by typing repeat motif) and repeat kind (perfect or imperfect). The flexibility is given to the user to limit the location of chromosome along with the number of markers required, range of GC %, range of base pairs and copy number.
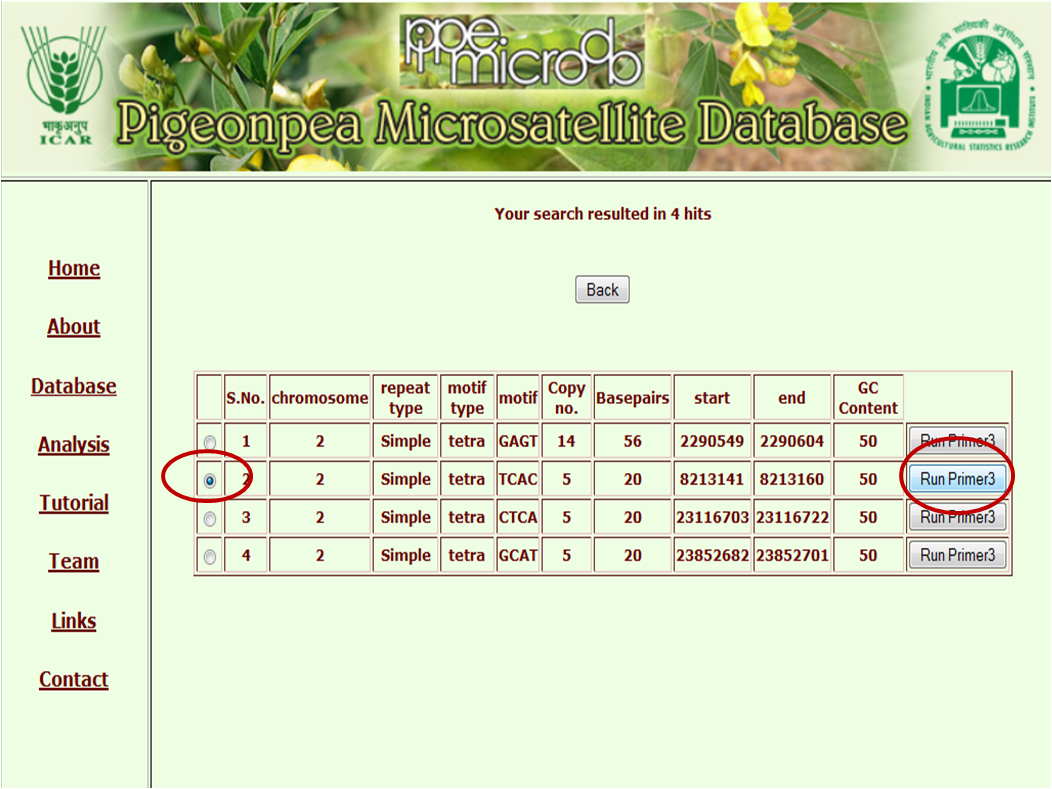
Based on the query combination, following output is achieved which shows the hit SSRs.
We get the following output, on performing the search based on multiple queries.
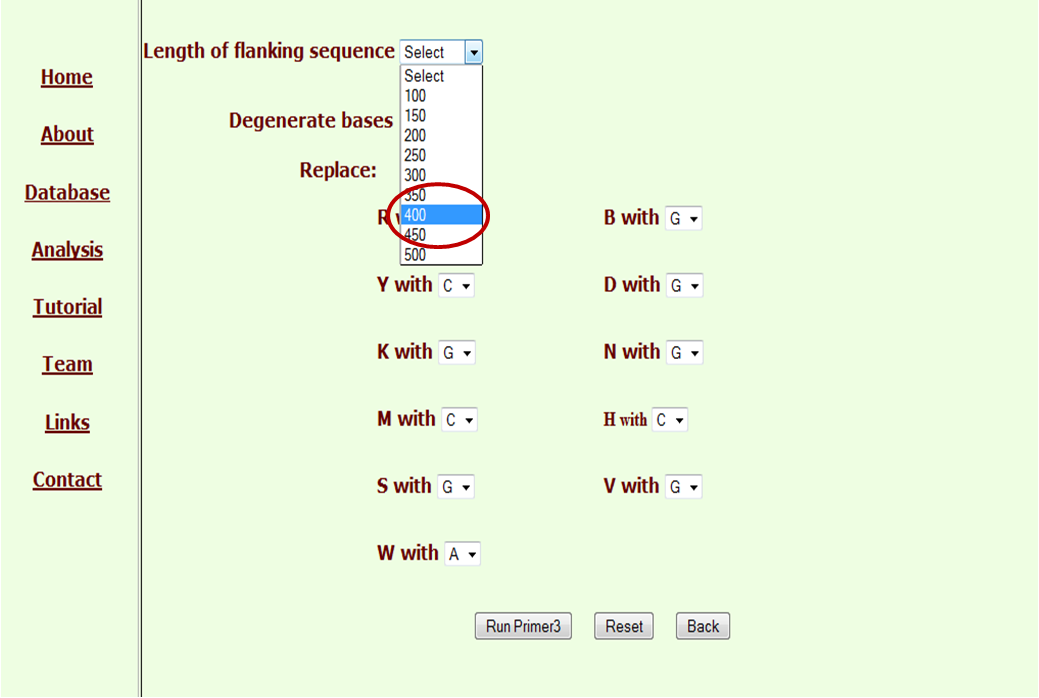
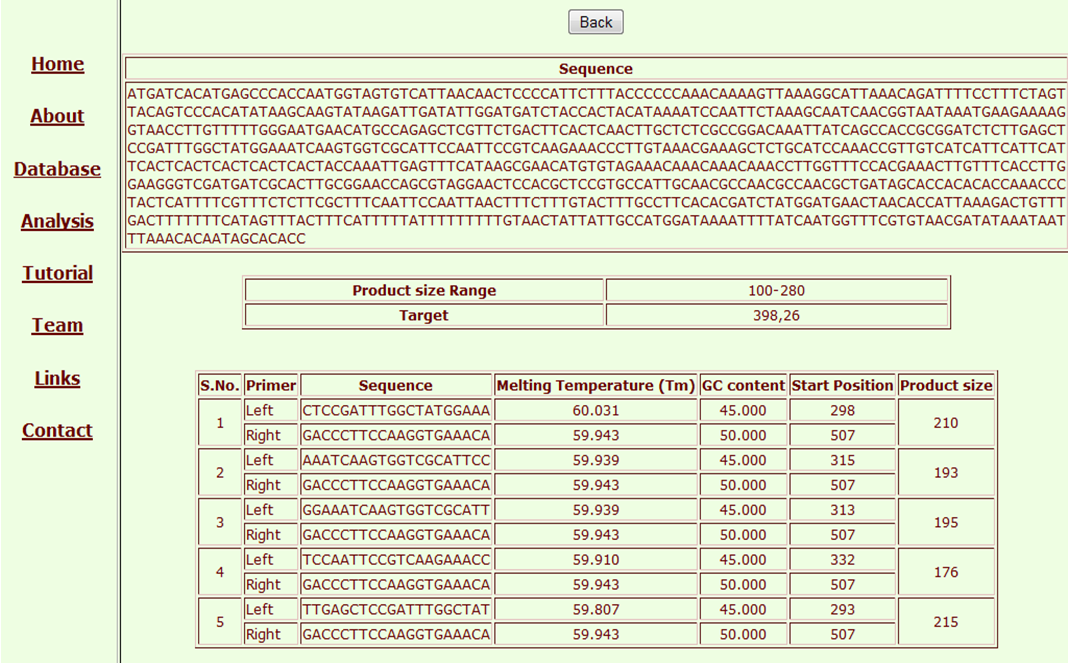
Now these may be selected one at a time to design a primer. On selection of Ssr and clicking Run Primer3 button, we get the option of either direct running Primer3 with default options or changing the bases for degenerate bases. This is novel flexibility not found in any of the microsatellite databases developed earlier. The flexibility of selecting the length of flankings is also provided.
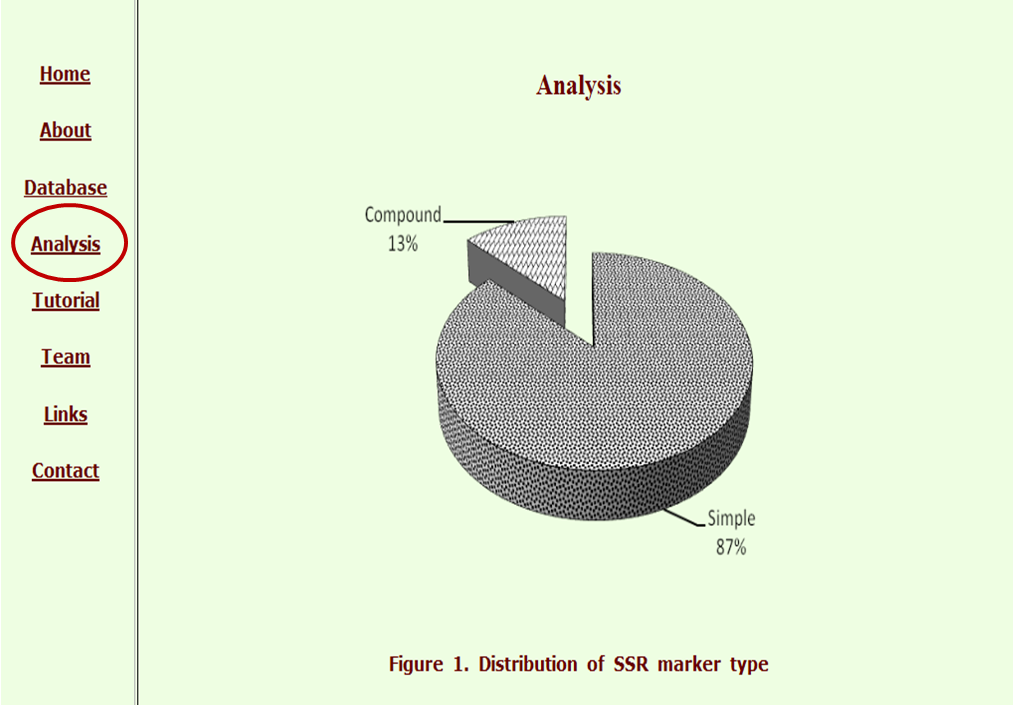
The “Analysis” tab gives the genome wide analysis of the data, shown graphically and in tabular form to get an idea of the genome trend in pigeonpea.
The “Link” tab contains the important links related to research in relevant fields and the genome resources.
“Team” and “Contact” tabs have the general information of the collaborators and the members involved in this project.
Glossary
- Allele: Any of two or more genes/DNA sequences that have the same relative position on homologous chromosomes and are responsible for alternative characteristics
- Base pair: Two complemetary nucleotides which form hydrogen bonds between the two antiparallel DNA strands.
- Chromosomes: A structural unit of inheritence; condensed DNA protein complex visible during cell division.
- Cloning: The process of making copies of a specific piece of DNA, usually a gene.
- Compound repeat: A compound microsatellite repeat is defined here as an occurrence of two or more microsatellites contiguously with intervening non-repeat sequence of <= 70 bp.
- Copy number: The number of times a particular motif has been tandemly repeated.
- Deletion: Loss of a base or more in the repeat track leaving one or more base of the repeat unit intact, e.g. (ATT)3TT(ATT)2.
- Dinucleotide repeat/ dimer : Tandem repeat of two nuclotides in the same order, e.g. ATATATATAT.
- DNA: Deoxyribonucleic acid. A molecule made out of two strands in antiparallel orientation that are held together by hydrogen bonds, each strand being made up of a sugar (deoxy ribose), a phosphate group and one of four bases (adenine, guanine, cytosine or thymine).
- Electrophoresis: The process in which DNA fragments can be separated according to size and electrical charge by applying an electric current to them, generally in a matrix of agarose gel or polyacrylamide gel.
- Exon: The region of a gene that encodes amino acids in a protein
- GC content : Guanine-Cytosine content of the genome of any given organism or any other piece of DNA or RNA
- Gene: One of the chromosomal units that transmit specific hereditary traits: a segment of the self- reproducing molecule, deoxyribonucleic acid
- Genetic code: The set of three nucleotides which transfer the information as a particular amino acid in protein during translation.
- Genetic map: A map of chromosomes showing the position of known genes and/or markers relative to each other, rather than as specific physical points on each chromosome
- Genetic Markers: Alleles of DNA polymorphisms, used as experimental tags to keep track of an individual, a tissue, a cell, a nucleus, a chromosome, or a gene. Stated another way, any character that acts as a signpost or signal of the presence OR location of a gene OR heredity characteristic in an individual of a population
- Genome: The total genetic complement of an organism, i.e. an organism's complete set of DNA sequences.
- Genotype: The actual alleles present in an individual; the genetic makeup of an organism.
- Heterozygous: Having two different alleles at the two corresponding loci on a pair of homologous chromosomes.
- Hexanucleotide repeat/ hexamer: Tandem repeat of six nucleotides in the same order, e.g. AATTTC AATTTC AATTTC AATTTC AATTTC.
- Homozygous: Having identical alleles at a single locus in the same individual.
- Imperfect repeat: A non-continuous repeat track, which is generated by a mutation (insertion/deletion/substitution) interrupting the continuity of the repeat unit., e.g. (ATT)3TT(ATT)2
- Insertion: The presence of an odd base(s) disturbing the continuity of the repeat or an additional similar or odd base(s) within the repeat unit e.g. (ATC)3G(ATC)3, (TGA)3TCGA(TGA)2.
- Intron: A noncoding sequence of DNA that is initially copied into RNA but is spliced out of the final RNA transcript.
- Linkage: The association of genes and/or markers that lie near each other on a chromosome. Linked genes and markers tend to be inherited together
- Locus: The position on a chromosome of a gene or a particular segment of DNA (marker).
- Microsatellite Family : A microsatellite family consists of all those microsatellites occurring in a genome, which possess highly matching flanking sequences, the stringency of the sequence match being: percentage match=95% and alignment length= 85%.
- Microsatellite: Tandem repeats of short simple DNA sequence, generally of 1-6 bases.
- Mononucleotide repeat/ monomer : Tandem repeat of single base, e.g. AAAAAAAAA.
- Motif: Recurring pattern in a DNA sequence.
- Multiplexing: The process of amplifying a number of different loci in a single polymerase chain reaction and the resolution of these loci either on polyacrylamide gel electrophoresis or agarose gel electrophoresis
- Mutation: Disruptions in the normal sequence of a DNA strand resulting in a varied sequence at the same position.
- Non-coding DNA : The segment of DNA that does not carry the information necessary to make an amino acid sequence.
- Nucleotide: One of the structural components, or building blocks, of DNA and RNA. A nucleotide consists of a base (one of five chemicals: adenine, thymine in DNA, uracil in RNA, guanine, and cytosine) plus a molecule of sugar and one of phosphoric acid
- Pentanucleotide repeat/ pentamer : Five nucleotides repeating in tandem to each other in the same order, e.g. AATTG AATTG AATTG AATTG.
- Phenotype: The visible characteristics of an organism, resultant of genotype and environment.
- Point mutation: A change in a single base pair
- Polymerase Chain Reaction: (PCR) A method of DNA analysis that exponentially amplifies a specific DNA sequence or region allowing rapid DNA analysis
- Polymorphic locus: A chromosomal region with two or more identifiable alleleic DNA sequences.
- Polymorphic: The occurrence of more than one form of individual in a single/multiple species within an interbreeding population
- Primer: A short oligonucleotide sequence used to amplify DNA sequences in a polymerase chain reaction
- Repeat element: Short stretches of DNA with the capacity to move between different points within a genome.
- Repeat kind: Nature of repeat sequence, e.g. Perfect Repeat Imperfect Repeat Compound Repeat
- Repeat type: Based on the number of nucleotides in a motif,repeat type is classified into mono,di,tri.tetra,penta,hexa.
- SSR: Simple sequence repeats (same as microsatellites). A repeated sequence of 1 to 6 bases.
- STR: Short tandem repeats (same as microsatellites). A repeated sequence of 1 to 6 bases.
- Substitution: A mutation, where one base is changed for another base.
- Tetranucleotide repeat/ tetramer : Tandem repeat of four nucleotides in the same order, e.g. ATAC ATAC ATAC ATAC.
- Threshold: A length limit, below which DNA sequence is not considered for the analysis, e.g. Threshold of 6 bases means, 6 base and above is considered but not 5 base.
- Transition: A mutation that consists of the replacement of one purine by another purine or one pyrimidine by another pyrimidine, e.g. Adenine to Guanine, Thymine to Cytosine.
- Transversion: A mutation that consists of the replacement of a purine by a pyrimidine or vice versa, e.g. Adenine to Thymine.
- Trinucleotide repeat/ trimer : Three nucleotides repeating in tandem to each other in the same order, e.g. TAA TAA TAA TA
.